| Citation: | Citation: XU AY, WANG TS, YANG T, et al. Constitution identification model in traditional Chinese medicine based on multiple features. Digital Chinese Medicine, 2024, 7(2): 108-119. DOI: 10.1016/j.dcmed.2024.09.002 |
To construct a precise model for identifying traditional Chinese medicine (TCM) constitutions, thereby offering optimized guidance for clinical diagnosis and treatment planning, and ultimately enhancing medical efficiency and treatment outcomes.
First, TCM full-body inspection data acquisition equipment was employed to collect full-body standing images of healthy people, from which the constitutions were labelled and defined in accordance with the Constitution in Chinese Medicine Questionnaire (CCMQ), and a dataset encompassing labelled constitutions was constructed. Second, heat-suppression valve (HSV) color space and improved local binary patterns (LBP) algorithm were leveraged for the extraction of features such as facial complexion and body shape. In addition, a dual-branch deep network was employed to collect deep features from the full-body standing images. Last, the random forest (RF) algorithm was utilized to learn the extracted multifeatures, which were subsequently employed to establish a TCM constitution identification model. Accuracy, precision, and F1 score were the three measures selected to assess the performance of the model.
It was found that the accuracy, precision, and F1 score of the proposed model based on multifeatures for identifying TCM constitutions were 0.842, 0.868, and 0.790, respectively. In comparison with the identification models that encompass a single feature, either a single facial complexion feature, a body shape feature, or deep features, the accuracy of the model that incorporating all the aforementioned features was elevated by 0.105, 0.105, and 0.079, the precision increased by 0.164, 0.164, and 0.211, and the F1 score rose by 0.071, 0.071, and 0.084, respectively.
The research findings affirmed the viability of the proposed model, which incorporated multifeatures, including the facial complexion feature, the body shape feature, and the deep feature. In addition, by employing the proposed model, the objectification and intelligence of identifying constitutions in TCM practices could be optimized.
The constitution in traditional Chinese medicine (TCM) reflects an individual’s distinct traits, offering insights into their response to external stimuli and susceptibility to health issues [1]. It serves as a comprehensive blueprint for one’s physical and mental constitution, shaped by TCM principles, enabling a deeper understanding of one’s health trajectory, disease prevention, and recovery processes [2]. Recent researches have shown a link between TCM constitution and various diseases, which has fostered the development of personalized healthcare strategies [3, 4]. By accurately identifying one’s TCM constitution, we can more effectively direct health interventions towards those who require them, ultimately improving their overall well-being.
The primary approach to identifying constitution in TCM involves completing the Constitution in Chinese Medicine Questionnaire (CCMQ) and subsequently applying statistical analysis methods to investigate and evaluate the results [5-7]. For example, YANG et al. [8] employed the CCMQ for the purpose of constitution identification, and further utilized statistical methods to explore the application of TCM constitution identification in health management and prevention in TCM treatment. Therefore, it is simple and easy to collect the constitution data of subjects by filling out the CCMQ. However, this approach has a problem of low efficiency.
In recent years, the advancement of computer vision and machine learning has led to the application of image processing techniques in TCM constitution identification [9]. Different TCM constitutions exhibit distinct tongue and facial features. For example, individuals with a balanced constitution may display a ruddy face and a pink tongue, while those with a phlegm-dampness constitution might exhibit much facial oil and a greasy tongue. ZHOU et al. [10] proposed a TCM constitution identification method based on tongue images, which used convolutional neural network (CNN), gray level co-occurrence matrix, and edge curve to extract features from tongue images, and employed support vector machine (SVM) classifier to classify various constitutions. YU [11] established a model for identifying TCM constitutions based on tongue images through multi-scale filtering CNN. LIANG [12] implemented an automatic identification system for TCM constitution based on facial complexion features incorporating skin color detection, SVM, and other technologies, subjecting facial images to analysis. YANG et al. [13] analyzed and compared the differences in facial complexion, lip color, and facial gloss in facial images of individuals with different constitutions, which provided important quantitative parameters for facial diagnosis in TCM constitution identification. Facial complexion and other information can be represented by heat-suppression valve (HSV) and other color spaces in images [14]. Therefore, image processing technology can realize automatic identification of TCM constitutions. With additional application of SVM, multi-scale filtering CNN, and other approaches, a high level of classification accuracy can be achieved in TCM constitution identification, thereby enhancing the reliability of the identification results.
Study has indicated that individuals with different constitutions exhibit significantly distinct physical traits [15]. For instance, individuals with a balanced constitution tend to possess a well-balanced and robust physique, whereas those with Qi deficiency constitution and Yang deficiency constitution may have soft and weak muscles. Body mass index (BMI) can serve as an indicator of physical characteristics to some extent. Many TCM constitution identification models have been constructed based on BMI. LU et al. [16] established a TCM constitution identification model by integrating BMI with features of tongue image. PAN et al. [17] extracted digital tongue image features and BMI to create an automatic TCM constitution identification model based on an artificial neural network and SVM. However, current related research falls short of fully exploiting the pertinent texture and structural characteristics present in the images to accurately represent body shape features.
With the continuous advancement of deep learning, the extraction of deep features using deep network models has been widely applied across various tasks [18, 19]. DONG et al. [20] proposed a strawberry pest classification method based on CNN to improve the classification accuracy. This method used the PyTorch deep learning framework to fine-tune AlexNet, which consists of five convolutional layers and three fully connected layers [21]. HE et al. [22] introduced residual connections and proposed a deep learning neural network architecture, ResNet, which swiftly trained extremely deep neural network models and prevented the gradient vanishing problem through cross-layer connections [23]. This network architecture performed well on tasks such as image recognition and object detection. GU et al. [24] proposed a deep learning model based on self-attention and visual converter structure to realize shape recognition. This model leveraged a Swin Transformer architecture and a comprehensive shape representation to enhance model performance [25]. The Swin Transformer employs a hierarchical structure and a design that are capable of processing and capturing deep features at multiple scales. This Swin Transformer-embedded model achieved an accuracy of 93.82% on the animal dataset, surpassing the visual geometry group (VGG) method by 3.8% [26]. Therefore, deep learning network models can not only automatically extract deep features, but also capture complex semantic information in images in a better manner than traditional methods. Deep features can effectively represent semantic information in images and enhance the performance of the model.
In the field of TCM, deep learning methods have been successfully employed for the intricate task of identifying TCM constitutions. ZHOU et al. [27] introduced an innovative approach that leveraged deep features extracted from tongue images to enhance the accuracy and efficiency of TCM constitution identification. This method improved the AlexNet network by incorporating an error-weight-based integration of features from each layer, thereby refining the network’s performance. Meanwhile, ZHOU et al. [28] proposed a TCM constitution identification model that utilized a dual-channel network, which combined tongue image features obtained manually with deep features to enhance the accuracy of TCM constitution classification. Therefore, the integration of features with multi-channel processing through deep learning networks can significantly improve the accuracy of identifying TCM constitutions. Unfortunately, current research endeavors in TCM constitution identification have yet to fully exploit the potential of deep learning networks for deeply mining and leveraging visual information from images.
However, most research on TCM constitution identification tends to focus solely on an individual aspect such as facial image or tongue image, resulting in limited outcomes confined to single TCM constitution identification models. This often fails to fully capture the comprehensive characteristics of an individual’s constitution. Therefore, this paper has surpassed existing deep learning methods by incorporating the attention mechanism, which has not only strategically directed the proposed network’s proficiency in extracting key features but also elevated the richness and accuracy of feature representation, leading to more comprehensive and insightful outcomes. In addition, this paper adopts a multi-feature approach, incorporating features such as facial complexion, body shape, and deep features extracted from full-body standing images using deep learning networks, with the aim of developing a TCM constitution identification model based on these multiple features.
In this paper, participants who voluntarily enrolled in the TCM constitution test were carefully selected from Nanjing University of Chinese Medicine, and the data collection period was from April 23 to May 10, 2023. Participants were eligible for the study if they did not have chronic diseases and signed an informed consent form; participants were excluded from the study if their clinical data of height, weight, or responses to questions were incomplete, or if they were unable to complete the questionnaire or respond to questions. The study protocol was approved by the Medical Ethics Committee of Affiliated Hospital of Nanjing University of Chinese Medicine (2023NL-255-01), in accordance with the guidelines of the Declaration of Helsinki of the World Medical Association.
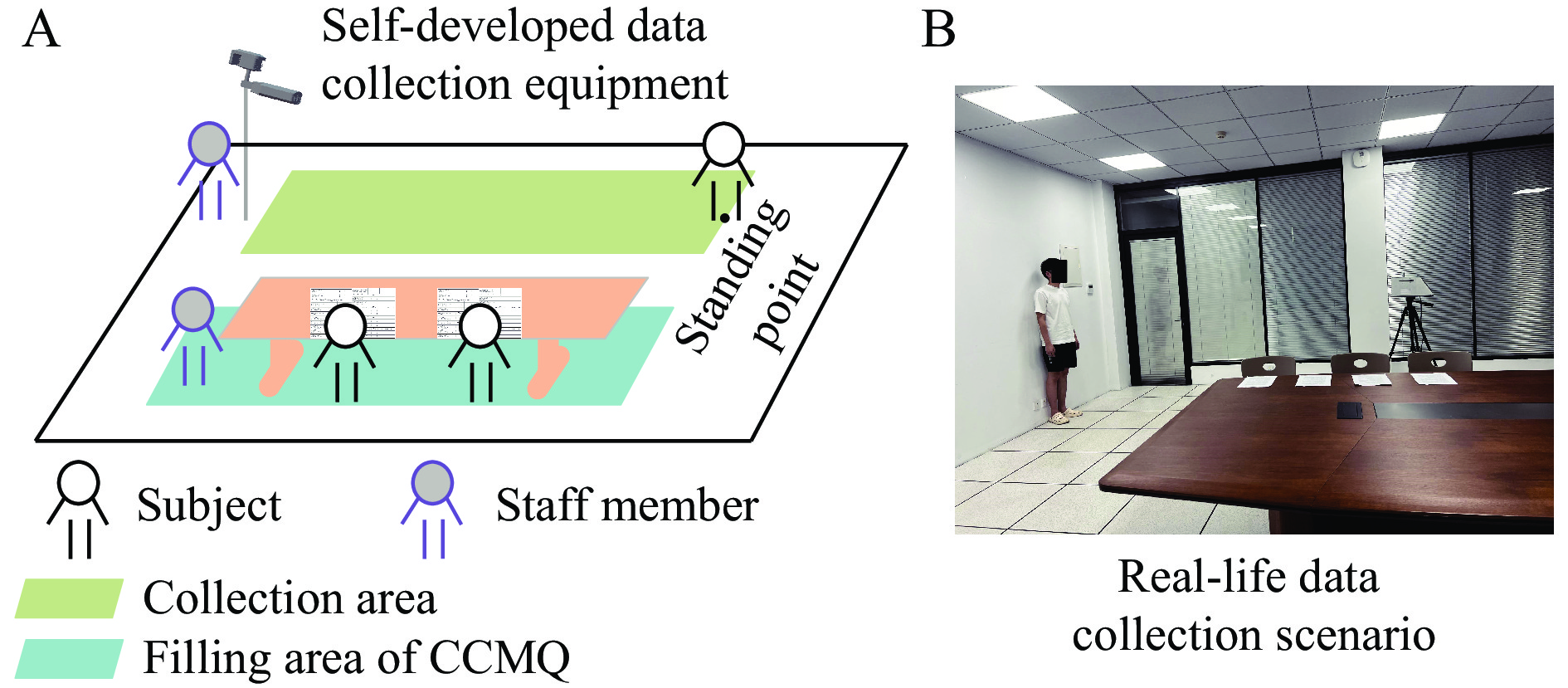
Figure 1 depicts the data acquisition scenario. The image acquisition area is designated as the green zone, with the self-developed data acquisition equipment strategically positioned to its left. This equipment incorporates a camera utilizing Femto technology, capable of capturing images with a resolution of 640 × 480 pixels, and is placed at a height of 1.5 m above the ground. Adjacent to the green area is the standing point, located 2 m away from the data acquisition equipment. The blue area is designed for completing the CCMQ, which was placed on the table for the subjects to fill out. The data acquisition process consisted of two steps. (i) The subject stood against the wall, waiting for the staff to operate the equipment to capture the full-body standing images. The purpose of collecting the full-body standing images was to extract subjects’ facial color features, body shape features, and deep features simultaneously. (ii) After their full-body images were captured, the subjects were asked to move to the blue area, where their TCM constitution types were determined and their heights and weights were measured, assisting in the construction of an image dataset with accurate constitution labels.
In this paper, the constitution of the subjects was assessed following the guidelines outlined in the “Classification and Determination of Constitution of TCM” published by the Chinese Association of Chinese Medicine in 2009 [29]. The subjects answered all the questions in the “Classification and Judgment Table of Chinese Medicine Constitution”, with each question rated on a 5-point scale. The subjects’ answers were used to calculate both the original score and the conversion score, and the constitution type was determined in accordance with predetermined criteria.
In this paper, a TCM constitution dataset was built with images and constitution data from 125 subjects. As illustrated in Figure 2, the dataset is comprised of 26 subjects with a balanced constitution and 99 with a biased constitution, encompassing various types such as Yang deficiency (21), Yin deficiency (20), Qi deficiency (16), dampness-heat (13), Qi stagnation (10), phlegm-dampness(9), inherited special constitutions (8), and blood stasis (2).
Figure 3 represents the proposed model framework in this paper, which was divided into four distinct stages: data acquisition, feature extraction, data partitioning, and model training. During the data acquisition stage, a self-developed data acquisition equipment was used to capture full-body standing images. Additionally, the TCM constitution was assessed using the CCMQ, and the height and weight were also measured to construct an image dataset annotated with constitution labels.
During the feature extraction phase, the facial color feature, body shape feature, deep feature, and BMI were all extracted and calculated as follows. (i) The YOLOv5 method was employed to identify the facial region in the full-body standing image. Subsequently, the red, green, and blue (RGB) facial image was converted to the HSV color space to extract the facial color feature
In the data partitioning phase, the dataset was split into a training set and a test set in a 7 : 3 ratio. Approaches such as SVM, multilayer perceptron (MLP), random forest (RF), and k-nearest neighbor (KNN) were employed to train the model. Subsequently, images in the test set were input into the four established models for identifying TCM constitutions. The performance of the models was verified with corresponding evaluation measures of accuracy, precision, and F1 value. Of the models, the one that exhibited the best performance was chosen as the final TCM constitution identification model.
The facial color feature serves as the foundation for identifying TCM constitutions [30]. It is a critical visual feature in the image that is insensitive to translation, scaling, or rotation, displaying a strong robustness. In this study, the facial color feature was extracted based on HSV color space (Figure 4).
Firstly, the object detection algorithm YOLOv5 [31] was utilized to analyze the full-body standing images so as to obtain the facial complexion, or in other terms, facial color. Figure 4 shows that the network architecture of YOLOv5 is divided into image input, backbone, neck, and detection head. The backbone, the core of the network, comprises modules such as Convolution Batchnorm SiLu (CBS), cross-stage partial network (CSP), and spatial pyramid pooling-fast (SPPF). The CBS module consists of convolutional layer, normalization, and activation function, facilitating effective down-sampling. The CSP module diminishes network complexity and computational demands, while SPPF dynamically adapts feature maps of varying sizes into uniform feature vectors. The neck is designed according to the feature pyramid and channel aggregation structures, enhancing feature integration. The detection header outputs the location, class, and confidence of the object.
Subsequently, the RGB image capturing the facial color feature was converted into HSV color space using Equations (1) − (3), where H represents hue, S represents saturation, and V represents brightness. Notably, the HSV color space offers a distinct advantage over the RGB model by decoupling image brightness from color information, leading to a color representation that more closely mirrors the human visual perception of color [32].
| Hij={arccos[(Rij−Gij)+(Rij−Bij)]2[(Rij−Gij)2+(Rij−Bij)+(Gij−Bij)]12,Bij≤Gij2π−arccos[(Rij−Gij)+(Rij−Bij)]2[(Rij−Gij)2+(Rij−Gij)+(Gij−Bij)]12,Bij>Gij | (1) |
Where
| Sij=max | (2) |
| {{V}}_{ij}=\frac{1}{255}\times \max\left({R}_{ij},{G}_{ij},{B}_{ij}\right) | (3) |
Next, the facial color features were extracted from the images. To realize this purpose, a mask was created to extract the skin region in the HSV color space, which was calculated as follows:
| {{\mathrm{Mask}}}_{ij} = \left\{ \begin{array}{*{20}{l}} 255 & {\mathrm{If}}\;{l}_{H}\le {H}_{ij}\le {u}_{H}\;\&\;{l}_{S}\le {S}_{ij}\le {u}_{S}\;\&\; {l}_{V}\le {V}_{ij}\le {u}_{V}\\ 0 & {\mathrm{Otherwise}}\end{array}\right. | (4) |
Where
Then, a bitwise AND operation was performed between the input RGB image and the mask to isolate the skin color region of the face image, and the calculation, formula is shown in Equation (5). Here,
| \begin{array}{l}{{\text {new}}{R}_{ij}=R}_{ij}\wedge {\text{mask}}_{ij}\\ {\text{new}}{G}_{ij}={G}_{ij}\wedge {\text{mask}}_{ij}\\ {\text {new}}{B}_{ij}={B}_{ij}\wedge {\text{mask}}_{ij}\end{array} | (5) |
Lastly, the pixel values within the skin color region were flattened into a one-dimensional vector, which was subsequently normalized. From this normalized vector, the 10 maximum pixel values were selected to represent the facial color feature.
People exhibit diverse body shapes depending on their constitutions, making the full-body body shape an essential basis for recognizing TCM constitutions. Given that LBP [33] is adept at capturing intricate details in images, this paper introduced a method for extracting the body shape feature using the LBP algorithm [34], as illustrated in Figure 5.
Firstly, the grayscale map of the RGB image was computed by Equation (6), where
| {g}_{ij}=0.3 \times {R}_{ij}+0.59\times {G}_{ij}+0.11\times {B}_{ij} | (6) |
Subsequently, the full-body standing image was analyzed using the LBP algorithm [35], which is:
| {{\mathrm{LBP}}}_{ij}=\sum _{k=0}^{P-1}s\left({g}_{k}-{g}_{ij}\right)\times {2}^{i} | (7) |
| {s}\left(x\right)=\left\{ \begin{array}{l}1,x\ge 0\\ 0,x < 0\end{array}\right. | (8) |
Where P represents the total number of sampling points placed on a circle centered around the pixel point, with P set to 8. For each sampling point k (where k = 0, 1, ..., P − 1),
Next, the LBP image was normalized using the grayscale map. The improved LBP map was obtained with the use of Equation (9), and
| {{\mathrm{ILBP}}}_{ij}=\frac{{{\mathrm{LBP}}}_{ij}}{{g}_{ij}} | (9) |
Subsequently, the feature map was generated using Equation (10), where
| {fm}_{ij}=\left\{\begin{array}{*{20}{l}} {{\mathrm{ILBP}}}_{ij} & {\mathrm{If}} \;0\le {{\mathrm{ILBP}}}_{ij}\le 9\\ 255 & {\mathrm{Otherwise}}\end{array}\right. | (10) |
Lastly, the shape feature vector was derived by computing the frequency histogram of the feature map. The Equation (11) for the shape feature vector is as follows:
| h\left(t\right)=\frac{c\left(t\right)}{{\sum }_{u=0}^{t}\,c\left(u\right)+\epsilon } | (11) |
Where t = 0, 1, ..., 9,
In this paper, the CResNet-ST method was introduced to extract deep features from full-body standing images (Figure 6). The method consists a dual-branch network architecture, with the upper half containing the Swin-T module, which is adept at capturing global features of the full-body standing image, and the lower half incorporating CResNet module, tailored for extracting local features from these images.
The attention mechanism assists the network in focusing on important regions within the image, enhancing its capability to extract essential features and thereby improving feature representation. The Swin-T module is a Swin Transformer network architecture that incorporates a self-attention mechanism. It is composed of five stages: Stage 1, 2, 3, 4, and 5. In Stage 1, the full-body standing image was divided into 4 × 4 size image patches using the Patch Partition layer. Stage 2 then transformed the input image patch into a low-dimensional feature space via the Linear Embedding layer to form the image patch
| \begin{split} {\widehat{Z}}^{l} =\; & {\text{MLP}}\left({\text{LN}}\left({\text{W\_MSA}}\left({\text{LN}}\left({Z}^{l}\right)\right)+{Z}^{l}\right)\right)+\\ & {\text{W\_MSA}}\left({\text{LN}}\left({Z}^{l}\right)\right)+{Z}^{l} \end{split} | (12) |
| \begin{split} {Z}^{l+1} =\; & {\text{MLP}}\left({\text{LN}}\left({\text{SW\_MSA}}\left({\text{LN}}\left({\widehat{Z}}^{l}\right)\right)+{\widehat{Z}}^{l}\right)\right)+\\&{\text{SW\_MSA}}\left({\text{LN}}\left({\widehat{Z}}^{l}\right)\right)+{\widehat{Z}}^{l}\end{split} | (13) |
Where, LN is the layer normalization operation that standardizes the input. W_MSA is a window self-attention module, MLP is a multi-layer perceptron module, and SW_MSA is a moving window self-attention module.
In Stage 3 − 5, the Patch Merging layer merged adjacent image patches, facilitating the consolidation of information. Following this, each consolidated patch underwent feature extraction and interaction within the Swin Transformer Block, collaboratively contributing to the construction of an enriched feature map. This sequence of Patch Merging and Transformer Block operations, consistently executed across all three stages, effectively processes the input image’s information, enhancing the model’s performance and representational capabilities while maintaining its lightweight architecture.
The CResNet module, an enhancement of the convolutional layer inspired by ResNet50 [25], innovatively incorporates the convolutional attention mechanism of CBAM [36]. This integration allows the network to sharpen its focus on significant features while minimizing disruptive noise interference. It is structured into five key stages: Stage 1 − 5. The CResNet module commences with Stage 1, which comprises two essential levels: Conv-CBAM and MaxPool. At the Conv-CBAM level, the full-body standing image underwent initial processing through a convolutional layer, subsequently refined by the application of the CBAM convolutional attention mechanism. The subsequent MaxPool layer then performed downsampling through the efficient maximum pooling method. The calculation formula for the CBAM is as follows.
| \begin{split} {F}' =\; & {\mathrm{Sigmod}}\; ({\mathrm{MLP}}({\mathrm{AvgPool}}(F))+ {\mathrm{MLP}}\;\\&({\mathrm{MaxPool}} (F))) \otimes F \end{split} | (14) |
| {F}''= {\mathrm{Sigmod}}\left({f}^{7\times 7}\left(\left[{\mathrm{AvgPool}}\left(F\right); {\mathrm{MaxPool}}\left(F\right)\right]\right)\right) \otimes{F}' | (15) |
Where F represents the feature map generated by the convolutional layer, which was then processed through the Sigmod activation function. The feature map underwent two parallel operations:
In Stage 2 − 5, the integration of three Conv-CBAM layers fostered robust feature propagation and seamless information flow, facilitated by the strategic deployment of residual blocks [37]. These residual blocks were designed to empower the network to delve deeper into learning complex feature representations, thereby enhancing the model's overall performance and efficiency. Within these stages, the primary function of these residual blocks was to aid the network in effectively capturing subtle variations and significant features within the data by linking input features to learned residuals. This mechanism has enhanced the model’s representational and generalization capabilities.
The heatmap intuitively displays the level of regional attention. Figure 8 shows the heatmap of deep features for different TCM constitutions, and the balanced constitution has a wider receptive field than that of the biased constitution. The prominence of a feature at its corresponding position within prior boxes correlates with the intensity of heat [38].
The computing platform environment employed in this paper was GeForce RTX 3080 GPU with 32 GB of RAM. The network construction, training, and testing were carried out using Python 3.7 and PyTorch 1.8.2 + cu111.
In this paper, a total of 125 subjects’ full-body standing images and constitution data were collected, and divided into a training dataset and a test dataset in a 7∶3 ratio. Regularization is a method used to avoid model overfitting and reduce generalization error [39]. Given that a small sample size affects the performance of the model, the Dropout regularization technique was introduced in the model training process to limit the complexity of the model and enhance its generalization ability. Accuracy, precision, and F1 score, three metrics generally for classification tasks, were selected for assessing the TCM constitution identification model as well. Accuracy is the percentage of samples that are correctly predicted by the model out of all the samples. Precision is the probability that all of the samples that are predicted to be positive are actually positive. F1 score is the harmonic mean of precision and recall. TP stands for true positive, TN for true negative, FP for false positive, and FN for false negative. The calculation formulas for these metrics are as follows.
| {\mathrm{Accuracy}}=\frac{{\mathrm{TP}}+{\mathrm{TN}}}{{\mathrm{TP}}+{\mathrm{TN}}+{\mathrm{FP}}+{\mathrm{FN}}} | (16) |
| {{\mathrm{Precision}}}=\frac{{\mathrm{TP}}}{{\mathrm{TP}}+{\mathrm{FP}}} | (17) |
| \mathrm{F}1= \;2\times \left(\frac{{\mathrm{TP}}}{{\mathrm{TP}}+{\mathrm{FP}}}\times \frac{{\mathrm{TP}}}{{\mathrm{TP}}+{\mathrm{FN}}}\right) \div \left(\frac{{\mathrm{TP}}}{{\mathrm{TP}}+{\mathrm{FP}}}+\frac{{\mathrm{TP}}}{{\mathrm{TP}}+{\mathrm{FN}}}\right) | (18) |
This paper has introduced four classification methods, which are RF [40], MLP, KNN, and SVM, for analysis and comparison to select the best one for TCM constitution identification. The comparison among the four classification models is shown in Table 1. It is evident that RF has outperformed MLP, KNN, and SVM in terms of accuracy. RF’s accuracy rate has increased by 0.053, 0.131, and 0.052 compared with MLP, KNN, and SVM, respectively. Moreover, F1 score of RF is 0.014 and 0.06 higher than that of MLP and KNN, respectively, while showing similar performance to SVM in terms of F1 score. Based on these results, the RF algorithm was chosen as the optimal classification model for TCM constitution identification.
| ID | Classifier | Accuracy | Precision | F1 score |
| 1 | RF | 0.842 | 0.868 | 0.789 |
| 2 | MLP | 0.789 | 0.766 | 0.775 |
| 3 | KNN | 0.711 | 0.755 | 0.729 |
| 4 | SVM | 0.790 | 0.790 | 0.790 |
Currently, there are many versions of YOLO object detection algorithm. YOLOv5 algorithm has excellent performance and is applied in many practical scenarios, based on which YOLOv8 was introduced in 2023. Different YOLO versions may have different effects on the experimental results. Therefore, this paper analyzed and compared the ability to detect objects in the full-body standing images with YOLOv5 and YOLOv8 (Figure 9). The results of YOLOv5 for object detection in full-body standing images are exactly the same as those of YOLOv8, with the accuracy, precision, and F1 score being 0.842, 0.868, and 0.790, respectively. The YOLO algorithms do not affect the extraction of the HSV color features, as they are capable of acquiring complete facial images.
To validate the effectiveness of the improved LBP in recognizing body shape features, this paper compared the identification performance of regular LBP and improved LBP with the same dataset. As illustrated in Figure 10, the improved LBP has demonstrated enhanced feature extraction capabilities compared with the regular LBP method. The accuracy, precision, and F1 score of the improved LBP increased by 0.053, 0.207, and 0.070, respectively, when compared with those of regular LBP. These results indicate that the improved LBP is more effective in extracting the body shape features.
To validate the effectiveness of the CResNet-ST introduced in this paper for identifying deep features, a performance comparison was made between CResNet-ST and Swin Transformer using the same dataset. As depicted in Figure 11, CResNet-ST showed improvements in accuracy, precision, and F1 score compared with those based on Swin Transformer features. Specifically, the accuracy, precision, and F1 score increased by 0.053, 0.102, and 0.015, respectively.
To further assess the capability of CResNet-ST in extracting deep features, a performance comparison was made again between CResNet-ST and Swin Transformer using the CIFAR-10 public dataset [41]. The network parameters were optimized by Adam optimizer [42] with a learning rate of
| Model | Training accuracy | Testing accuracy |
| Swin Transformer | 0.963 | 0.813 |
| CResNet-ST | 0.988 | 0.926 |
To validate the efficiency of the models based on multi-features, the accuracy, precision, and F1 score of these models were assessed utilizing the same dataset as shown in Table 3. In comparison with the identification models that encompass a single feature, either a facial complexion feature, a body shape feature, or deep features, the accuracy of the model that incorporates all the aforementioned features was elevated by 0.105, 0.105, and 0.079, the precision increased by 0.164, 0.164, and 0.211, and the F1 score rose by 0.071, 0.071, and 0.084, respectively. The accuracy of the proposed model based on multi-features reached 0.842. In many studies [16, 17], BMI is an important and effective feature that reflects an individual’s body shape. Therefore, a model on the basis of BMI, integrating body shape features, facial color features, and deep features, was built. The F1 score of the introduced model in this paper is higher by 0.010 compared with that of a single BMI feature model. The accuracy and precision increased by 0.026 and 0.062, respectively. Generally, the proposed multi-features-based model outperformed the model encompassing only one feature.
| ID | Feature | Accuracy | Precision | F1 score |
| 1 | BMI | 0.816 | 0.806 | 0.810 |
| 2 | Body shape feature | 0.737 | 0.704 | 0.719 |
| 3 | Facial color feature | 0.737 | 0.704 | 0.719 |
| 4 | Deep feature | 0.763 | 0.657 | 0.706 |
| 5 | BMI, body shape feature, and facial color feature |
0.816 | 0.666 | 0.733 |
| 6 | BMI, body shape feature, and deep feature |
0.790 | 0.662 | 0.720 |
| 7 | BMI, facial color feature, and deep feature |
0.790 | 0.662 | 0.720 |
| 8 | Body shape feature, facial color feature, and deep feature | 0.737 | 0.704 | 0.719 |
| 9 | BMI, body shape feature, facial color feature, and deep feature | 0.842 | 0.868 | 0.790 |
To analyze the performance of the TCM constitution identification model proposed in this study, its accuracy, precision, and F1 score for identifying balanced, Yindeficient, and Yang deficient constitutions were evaluated. In addition, the performance of the model was compared with that of other models. Table 4 illustrates that the multi-features-based constitution identification model achieved an accuracy of 0.714, a precision of 0.793, and an F1 score of 0.721 for recognizing Balanced constitution, Yin deficient constitution and Yang deficient constitution. When compared with the TCM constitution identification model based on human skeleton features [43], the accuracy, precision, and F1 score of this model were improved by 0.143, 0.105, and 0.143, respectively. Compared with the TCM constitution identification model based on CNN [10], the accuracy, precision and, F1 score of this model were improved by 0.143, 0.245, and 0.178, respectively. These results have demonstrated that the TCM constitution identification model utilizing multi-features outperformed other models in terms of identifying TCM constitutions.
The comparison between the actual constitution and the predicted constitution by the proposed TCM constitution identification model using the testing set is shown in Figure 13. Figure 13A is well-proportioned and robust, which leans towards balanced constitution, and Figure 13B is thin, which is leaning towards Yin deficiency constitution. The TCM constitution identification model proposed in this paper was able to correctly identify the differences between Figure 13A and 13B.
Constructing a precise model for identifying TCM constitutions can provide better guidance for formulating clinical diagnoses and treatment plans, thereby enhancing medical efficiency and treatment outcomes. Traditional methods [6], such as filling out the CCMQ, have the problem of low efficiency. With the development of computer vision, constitution identification models based on tongue and facial images have been widely studied. Research indicates that TCM constitution is closely associated with facial complexion and body shape [16]. However, most current studies focus on extracting the features from certain parts of the human body, which cannot represent the comprehensive features related to constitution. In this paper, a TCM constitution recognition model based on multi-features is proposed. This model extracts facial complexion, body shape, and deep features from full-body standing images to improve the accuracy of constitution identification.
The study results demonstrated that the predictive accuracy of the model based on multi-features was higher by 0.105, 0.105, 0.079, and 0.026 than that of the single feature based model, including the single facial complexion feature model, body shape feature model, deep feature model, and BMI model, respectively. Meanwhile, the proposed model achieved a high accuracy of 0.842, indicating good performance. The study results also confirmed the feasibility of the proposed multi-feature-based model. Nonetheless, the proposed model in the study is expected to further improve the objectivity and intelligence of constitution identification in TCM practices.
However, the sample size in this study was limited, including only 125 subjects. Given that a small sample size could affect the performance of the model, this paper introduced the Dropout regularization technique in the training process to limit the complexity of the model and enhance its generalization ability. Additionally, recognizing that the potential for error stems from the reliance on a single data source, it’s necessary to carry out multi-level and multi-regional data validation for the assessment of the model’s applicability across diverse environments in future studies.
In this study, a TCM constitution identification model based on multi-features, including facial complexion, body shape feature, deep feature, and BMI, was proposed. This study introduced the extraction of facial complexion features based on the HSV color space, the enhancement of the LBP algorithm for extracting body shape features, and the dual-branch CResNet-ST method for extracting deep features from the full-body standing images. TCM constitution identification requires comprehensive information about a person to analyze and judge. In future research, the data of different people will be collected to improve the applicability of the model in different environments. At the same time, tongue, hand, and other data will be included to further improve the TCM constitution identification model, enhancing its accuracy and reliability.
National Key Research and Development Program of China (2022YFC3502302), National Natural Science Foundation of China (82074580), and Graduate Research Innovation Program of Jiangsu Province (KYCX23_2078).
Competing Interests: The authors declare no conflict of interest
| [1] |
SHEN YQ, LIAO SH, SHEN Y, et al. Research progress of traditional Chinese medicine constitution based on primary disease of chronic kidney disease. Chinese Journal of Ethnomedicine and Ethnopharmacy, 2024, 33(7): 57–61. doi: 10.3969/j.issn.1007-8517.2024.07.zgmzmjyyzz202407013
|
| [2] |
FENG MY, OUYANG MH, PAN ZS, et al. Study on the characteristics of traditional Chinese medicine constitution and the findings of meridian detection in overweight and obese population. Journal of Guangzhou University of Traditional Chinese Medicine, 2023, 40(9): 2153–2159. doi: 10.13359/j.cnki.gzxbtcm.2023.09.005
|
| [3] |
ZHENG TT, ZHANG BX, NI PP, et al. Research progress on the correlation between lung cancer and TCM constitution identification. China & Foreign Medical Treatment, 2023, 42(11): 189–194. doi: 10.16662/j.cnki.1674-0742.2023.11.189
|
| [4] |
LING J, MO HF, JIANG ZX, et al. Progress of traditional Chinese medicine constitution identification on influence of hypertensive pathogenesis in community. Clinical Journal of Traditional Chinese Medicine, 2022, 34(12): 2397–2401. doi: 10.16448/j.cjtcm.2022.1245
|
| [5] |
LONG LQ, ZHU YB, CHEN PP. Analysis of TCM constitution factors of public acute stress reaction during COVID-19 epidemic based on sample data of 1102 cases. Journal of Beijing University of Traditional Chinese Medicine, 2022, 45(12): 1242–1248. doi: 10.3969/j.issn.1006-2157.2022.12.008
|
| [6] |
QI XZ, YAN MQ. Study on the relation between sleep status and the constitution of traditional Chinese medicine among naval special service personnel. Journal of Navy Medicine, 2023, 44(1): 7–11. doi: 10.3969/j.issn.1009-0754.2023.01.002
|
| [7] |
YAN L, ZHOU ZJ, SONG YH, et al. Study on the identification model of traditional Chinese medicine constitutions based on ML-KNN multi-label learning. Modernization of Traditional Chinese Medicine and Materia Medica-World Science and Technology, 2020, 22(10): 3558–3562. doi: 10.11842/wst.20200319009
|
| [8] |
YANG LL, XUE Y, WANG YP, et al. Application of TCM constitution identification in health management of preventive treatment of disease in TCM. China Journal of Traditional Chinese Medicine and Pharmacy, 2018, 33(10): 4595–4598. doi: CNKI:SUN:BXYY.0.2018-10-091
|
| [9] |
ZHOU SJ, TU YQ, HUANG ZP, et al. Study on the auxiliary identification of TCM constitution based on tongue image feature extraction. Lishizhen Medicine and Materia Medica Research, 2013, 24(11): 2734–2735. doi: 10.3969/j.issn.1008-0805.2013.11.071
|
| [10] |
ZHOU H, HU G, ZHANG X. Constitution identification of tongue image based on CNN. 2018 11th International Congress on Image and Signal Processing. BioMedical Engineering and Informatics (CISP-BMEI), 2018: 1-5.
|
| [11] |
YU ZT. Research on traditional Chinese medicine constitution identification algorithm based on neural network. Nanjing: Nanjing University of Science & Technology, 2021.
|
| [12] |
LIANG YM. Research on automatic identification system of TCM constitution based on facial image feature. Beijing: Beijing University of Technology, 2016.
|
| [13] |
YANG S, XU Y, GUAN X, et al. Analysis and research on characteristics of face diagnosis image of people with Yin-deficiency and Yang-deficiency constitution. Journal of Basic Chinese Medicine, 2021, 27(7): 1141–1144. doi: 10.19945/j.cnki.issn.1006-3250.2021.07.025
|
| [14] |
PRAKASH SR, SINGH PN. Background region based face orientation prediction through HSV skin color model and K-Means clustering. International Journal of Information Technology, 2023, 15(3): 1275–1288. doi: 10.1007/s41870-023-01174-1
|
| [15] |
YANG XG, LI XZ, REN Y, et al. Classification of traditional Chinese medicine constitution and questionnaire: application and research. Chinese Journal of Integrated Traditional and Western Medicine, 2017, 37(8): 1003–1007. doi: 10.7661/j.cjim.20170426.127
|
| [16] |
LU GL, HUANG YS, ZHANG Q, et al. The study of auxiliary TCM constitution identification model based on tongue image and physical features. Lishizhen Medicine and Materia Medica Research, 2019, 30(1): 244–246. doi: 10.3969/j.issn.1008-0805.2019.01.085
|
| [17] |
PAN SX, LIN Y, ZHOU SJ, et al. The study of traditional Chinese medical constitution identification models based on artificial neural network and support vector machine. Modernization of Traditional Chinese Medicine and Materia Medica-World Science and Technology, 2020, 22(4): 1341–1347. doi: 10.11842/wst.20190417003
|
| [18] |
PUSHPA BR, ADARSH A, SHREE HA. Plant disease detection and classification using deep learning model. 2021 Third International Conference on Inventive Research in Computing Applications (ICIRCA), 2021: 1285-1291.
|
| [19] |
ATILA Ü, UÇAR M, AKYOL K, et al. Plant leaf disease classification using EfficientNet deep learning model. Ecological Informatics, 2021, 61: 101182. doi: 10.1016/j.ecoinf.2020.101182
|
| [20] |
DONG C, ZHANG ZW, YUE J, et al. Classification of strawberry diseases and pests by improved AlexNet deep learning networks. 2021 13th International Conference on Advanced Computational Intelligence (ICACI). IEEE, 2021: 359-364.
|
| [21] |
KRIZHEVSKY A, SUTSKEVER I, HINTON GE. Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems, 2012: 25.
|
| [22] |
HE KM, ZHANG XY, REN SQ, et al. Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2016: 770-778.
|
| [23] |
XU WN, FU YL, ZHU DM. ResNet and its application to medical image processing: research progress and challenges. Computer Methods and Programs in Biomedicine, 2023, 240: 107660. doi: 10.1016/j.cmpb.2023.107660
|
| [24] |
GU T, MIN R. A swin transformer based framework for shape recognition. Proceedings of the 2022 14th International Conference on Machine Learning and Computing, 2022: 388-393.
|
| [25] |
LIU Z, LIN YT, CAO Y, et al. Swin transformer: hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF International Conference on Computer Vision, 2021: 10012-10022.
|
| [26] |
SIMONYAN K, ZISSERMAN A. Very deep convolutional networks for large-scale image recognition. arXiv, 2014. doi: 10.48550/arXiv.1409.1556.
|
| [27] |
ZHOU H, HU GQ, ZHANG XF. Preliminary study on traditional Chinese medicine constitution classification method based on tongue image feature fusion. Beijing Biomedical Engineering, 2020, 39(3): 221–226. doi: 10.3969/j.issn.1002-3208.2020.03.001
|
| [28] |
ZHOU H, ZHANG XF, HU GQ, et al. Preliminary study on classification of traditional Chinese medicine constitution types based on feature fusion of dual-channel network. Proceedings of the 13th National Academic Symposium of the Diagnostic Professional Committee of the China Association of Chinese and Western Medicine. China Association of Chinese and Western Medicine, 2019: 19.
|
| [29] |
China Association of Chinese Medicine. Classification and determination of TCM constitution. Beijing: China Traditional Chinese Medicine Press, 2009.
|
| [30] |
SUN YY, XIE W, JIA QN, et al. Study on the characteristics of infrared thermal imaging of hand and face in the people with constitution of Yang-deficiency or Yin-deficiency. Modern Journal of Integrated Traditional Chinese and Western Medicine, 2020, 29(31): 3427–3431, 3450. doi: 10.3969/j.issn.1008-8849.2020.31.002
|
| [31] |
ZHANG Y, GUO Z, WU J, et al. Real-time vehicle detection based on improved YOLO v5. Sustainability, 2022, 14(19): 12274. doi: 10.3390/su141912274
|
| [32] |
ZHANG YZ, WANG Y, CHENG P, et al. Identification method of gardeniae fructus praeparatus processing degree based on color features and support vector machine. China Journal of Traditional Chinese Medicine and Pharmacy, 2023, 38(8): 3768–3772.
|
| [33] |
HU YC, LI QS. Face recognition method based on haar-like T and LBP features. Automation & Instrumentation, 2023, 38(10): 52–56, 61. doi: 10.19557/j.cnki.1001-9944.2023.10.012
|
| [34] |
OJALA T, PIETIKAINEN M, MAENPAA T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2002, 24(7): 971–987. doi: 10.1109/TPAMI.2002.1017623
|
| [35] |
HAO ZY. Image texture feature extraction method and application based on LBP. Taiyuan: Taiyuan University of Science & Technology, 2023.
|
| [36] |
WOO S, PARK J, LEE JY, et al. CBAM: convolutional block attention module. Proceedings of the European Conference on Computer Vision (ECCV), 2018: 3-19.
|
| [37] |
LIU YB, GAO WB, HE P, et al. Apple phenological period identification in natural environment based on improved ResNet50 model. Smart Agriculture, 2023, 5(2): 13–22. doi: 10.12133/j.smartag.SA202304009
|
| [38] |
HUANG L, ZHANG H, HU XL, et al. Recognition and localization method for pepper clusters in complex environments based on improved YOLOv5. Transactions of the Chinese Society for Agricultural Machinery, 2024, 55(3): 243–251. doi: 10.6041/j.issn.1000-1298.2024.03.024
|
| [39] |
CHEN K, WANG AZ. Survey on regularization methods for convolutional neural network. Application Research of Computers, 2024, 41(4): 961–969. doi: 10.19734/j.issn.1001-3695.2023.06.0347
|
| [40] |
BREIMAN L. Random forests. Machine Learning, 2001, 45(1): 5–32. doi: 10.1023/A:1010933404324
|
| [41] |
RECHT B, ROELOFS R, SCHMIDT L, et al. Do CIFAR-10 classifiers generalize to CIFAR-10, arXiv, 2018. doi: 10.48550/arXiv.1806.00451
|
| [42] |
LOSHCHILOV I, HUTTER F. Decoupled weight decay regularization. arXiv, 2017. doi: 10.48550/arXiv.1711.05101.
|
| [43] |
WANG ZY, LI HY, ZHOU ZJ, et al. Research of identifying traditional Chinese medicine constitution model based on human skeleton feature. Chinese Archives of Traditional Chinese Medicine, 2024, 42(8): 37−40, 266−267.
|
| ID | Classifier | Accuracy | Precision | F1 score |
| 1 | RF | 0.842 | 0.868 | 0.789 |
| 2 | MLP | 0.789 | 0.766 | 0.775 |
| 3 | KNN | 0.711 | 0.755 | 0.729 |
| 4 | SVM | 0.790 | 0.790 | 0.790 |
| Model | Training accuracy | Testing accuracy |
| Swin Transformer | 0.963 | 0.813 |
| CResNet-ST | 0.988 | 0.926 |
| ID | Feature | Accuracy | Precision | F1 score |
| 1 | BMI | 0.816 | 0.806 | 0.810 |
| 2 | Body shape feature | 0.737 | 0.704 | 0.719 |
| 3 | Facial color feature | 0.737 | 0.704 | 0.719 |
| 4 | Deep feature | 0.763 | 0.657 | 0.706 |
| 5 | BMI, body shape feature, and facial color feature |
0.816 | 0.666 | 0.733 |
| 6 | BMI, body shape feature, and deep feature |
0.790 | 0.662 | 0.720 |
| 7 | BMI, facial color feature, and deep feature |
0.790 | 0.662 | 0.720 |
| 8 | Body shape feature, facial color feature, and deep feature | 0.737 | 0.704 | 0.719 |
| 9 | BMI, body shape feature, facial color feature, and deep feature | 0.842 | 0.868 | 0.790 |